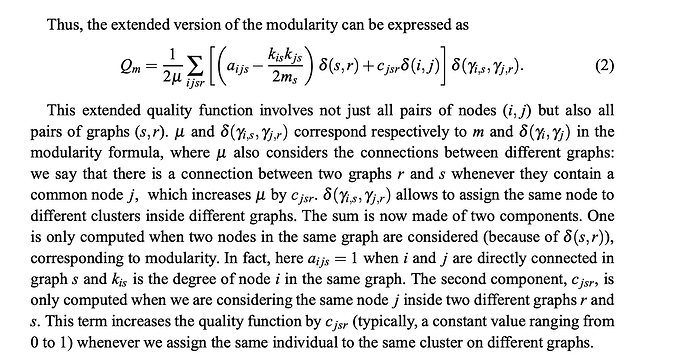
I am trying to implement the following extended modularity metric on multi-layer graphs as presented by the following paper: https://arxiv.org/pdf/1501.01676.
Below is my implementation using graph-tool of the multi-layer network defined in figure 6 for which modularity values were reported for varying values of the parameter c (omega in my code).
I have defined this network by the below code:
def bothorel_mln():
# isolated node 6 -> represented as self loop with weight 0
layer0 = [(1, 2, 1),
(1, 3, 1),
(2, 3, 1),
(4, 7, 1),
(5, 7, 1),
(6, 6, 0)]
# added one extra node 8 in this layer
layer1 = [(1, 2, 1),
(1, 3, 1),
(2, 3, 1),
(4, 5, 1),
(4, 6, 1),
(5, 8, 1),
(6, 7, 1),
(6, 8, 1),
(7, 8, 1)]
layer2 = [(1, 2, 1),
(1, 3, 1),
(2, 3, 1),
(4, 5, 1),
(4, 8, 1),
(5, 7, 1),
(6, 6, 0),
(7, 8, 1)]
intralayer_edges = [layer0, layer1, layer2]
# Create multi layer graph with no interlayer edges
G = multilayer_graph_extended(intralayer_edges, {}, directed=False)
# Layer-aware community assignment per (original ID, layer)
communities = {
(1,0): 0, (1, 1): 0, (1, 2): 0,
(2, 0): 0, (2, 1): 0, (2, 2): 0,
(3, 0): 0, (3, 1): 0, (3, 2): 0,
(4, 0): 1, (4, 1): 1, (4, 2): 1,
(5, 0): 1, (5, 1): 1, (5, 2): 1,
(6, 0): 1, (6, 1): 1, (6, 2): 1,
(7, 0): 1, (7, 1): 1, (7, 2): 1,
(8,1): 1, (8,2): 1
}
# Create a new vertex property for communities
G.vp["community"] = G.new_vertex_property("int")
# Assign per (node, layer)
for v in G.vertices():
node_layer = (G.vp["original_id"][v], G.vp["layer_id"][v]) # Identify node-layer pair
if node_layer in communities:
G.vp["community"][v] = communities[node_layer]
return G
And my extended modularity function is based on the classical modularity function of a single graph which I implemented below:
Classical modularity on a single layer graph (verified and tested):
def graph_matrix_modularity(graph: gt.Graph) -> float:
"""
Computes the modularity of the input graph using a matrix-based approach.
Args:
graph (gt.Graph): The graph-tool Graph object.
Returns:
float: The modularity score of the graph.
"""
if "community" not in graph.vp:
raise ValueError("Community assignment missing. Ensure 'community' vertex property is set.")
# Set high precision for Decimal calculations
getcontext().prec = 30
# Convert to adjacency matrix (numpy dense format)
A = graph_tool.spectral.adjacency(graph, weight=graph.ep["weight"]).toarray()
# print(A)
# Compute degree vector (sum of edge weights per node)
k = A.sum(axis=1) # Degree sum per node
m = Decimal(k.sum()) / 2 # Total edge weight (undirected)
# Retrieve community assignments as an array
communities = np.array([graph.vp["community"][v] for v in graph.vertices()])
# Compute expected weight matrix P_ij = (k_i * k_j) / (2m)
P = np.outer(k, k) / (2 * float(m))
# Compute community mask: M_ij = 1 if nodes i, j are in the same community, else 0
M = (communities[:, None] == communities[None, :]).astype(int)
# Compute modularity using matrix operations
modularity_matrix = (A - P) * M # Element-wise multiplication
modularity = Decimal(modularity_matrix.sum()) / (2 * m)
# Round precision
modularity = modularity.quantize(Decimal("0.000001"))
return float(modularity)
Extended modularity (on which I am having some issues)
def mln_matrix_extended_modularity(graph: gt.Graph, omega: float = 1.0) -> float:
"""
Computes the extended modularity for a multi-layer or multi-slice graph using matrix operations.
Args:
graph (gt.Graph): The multilayer/multislice graph object.
gamma (float): Resolution parameter for intra-layer modularity.
omega (float): Coupling strength for inter-layer edges.
Returns:
float: The extended modularity score.
"""
if "community" not in graph.vp:
raise ValueError("Community assignment missing. Ensure 'community' vertex property is set.")
num_layers = graph.gp["num_layers"]
# Step 1: Compute the intra-layer modularity (s = r)
intra_layer_modularity = 0.0
for layer in range(num_layers):
# Extract subgraph for the current layer
subgraph = graph_tool.GraphView(graph, vfilt=lambda v: graph.vp["layer_id"][v] == layer)
# Compute modularity for this layer
subgraph_modularity = single.graph_matrix_modularity(subgraph)
print(f"Modularity of layer {layer}: {subgraph_modularity}")
# Sum total weight for normalization later
layer_weight = sum(graph.ep["weight"][e] for e in graph.edges() if graph.ep["source_layer"][e] == layer and not graph.ep["is_interlayer"][e])
# Intra-layer modularity with scaling factor removed (divison by 2m)
intra_layer_modularity += subgraph_modularity * (2 * layer_weight)
# Step 2: Compute inter-layer contribution
inter_layer_contribution = 0.0
node_replicas = defaultdict(set)
# Counting contributions based on layer-layer relationships rather than a node overall's presence
for v in graph.vertices():
node_id = graph.vp["original_id"][v]
layer_id = graph.vp["layer_id"][v]
node_replicas[node_id].add(layer_id)
# Iterate over each unique node in the multilayer graph
for node, layers in node_replicas.items():
layer_list = sorted(layers) # Ensure a consistent order of layers
# Compare all layer pairs for this node, including duplicates
for s in layer_list:
for r in layer_list: # Instead of ensuring r > s, we allow full pairwise comparison
if s != r: # Avoid self-comparisons within the same layer
# Get the node's vertex in each layer
v_s = next((v for v in graph.vertices() if graph.vp["original_id"][v] == node and graph.vp["layer_id"][v] == s), None)
v_r = next((v for v in graph.vertices() if graph.vp["original_id"][v] == node and graph.vp["layer_id"][v] == r), None)
# Ensure the node exists in both layers before proceeding
if v_s is not None and v_r is not None:
# Check if the node is in the same community in both layers
if graph.vp["community"][v_s] == graph.vp["community"][v_r]:
inter_layer_contribution += omega
# print(f"Node {node} appears in Layer {s} and Layer {r} (double counted): +{omega} (Community {graph.vp['community'][v_s]})")
# Step 3: Compute final total scaling factor
M_total = compute_extended_modularity_total_scaling_factor(graph, omega)
print("\n")
print(f"Total scaling factor (M): {M_total}")
print(f"Total intra layer modularity (s=r term): {intra_layer_modularity}")
print(f"Total inter layer contribution (s≠r term): {inter_layer_contribution}")
# Step 4: Normalize modularity
extended_modularity = (intra_layer_modularity + inter_layer_contribution) / (2 * M_total)
# Round precision
extended_modularity = round(extended_modularity, 6)
return extended_modularity
For c = 0, I am getting a modularity of 0.468027 which is close to the value of 0.48 reported in the paper. However when c = 1, the gap in the value is much larger. I obtain 0.740199 vs 0.68 reported in the paper.
Since I am 100% sure that my single layer modularities are computed correctly, the error must lie either within the scaling factor (M_total) or the sum of inter layer contributions. Currently, in my code, my mu parameter (M_total) contains the number of all intra-layer edges in the network (including duplicate edges across layers) and the number of shared ndoes across pairs of layers (where each pair is considered once so in this case (layer1, layer 2), (layer2, layer3) and (layer1, layer3) where in each case there are 7 + 8 + 7 = 22 shared nodes respectively). However in the second term of the modularity sum where the parameter c appears, I am counting the appearance of a node in the same cluster across the same pair of layers counted twice so I would consider the j in layer (s,r) and (r,s). If I consider pairs uniquely, the modularity value is way too low.